With substantial resources devoted to clinical trials and drug development in immuno-oncology, big data analysis has the potential to uncover important mechanisms underlying immunotherapy and, in turn, offer novel insights for their clinical management in immunotherapy.
Harnessing big data for precision oncology
In the Han Lab, we aim to utilize cutting-edge techniques in computational biology, RNA biology, and systems biology to identify novel prognostic and diagnostic biomarkers and to develop innovative therapeutic strategies.

Meet the PI: Leng Han, PhD
- Cutting-edge techniques in systems biology to understand the molecular mechanisms of complex diseases
- David Brown Professor of Genomic Medicine
- Professor of Biostatistics & Health Data Science
MSWuhan University
PhDChinese Academy of Sciences
PostdocStanford University, MD Anderson Cancer Center
What’s happening in the Han Lab?
Data-driven, large-scale multi-omics
Recent advances in genomic characterization technologies and the explosive genomic information related to disease have accelerated the convergence of discovery science with clinical medicine.
Successful translations of genomics into therapeutics and diagnostics reinforce the potential for precision medicine. Analyses of multi-dimensional data provide unique opportunities to understand biological processes and signaling pathways.
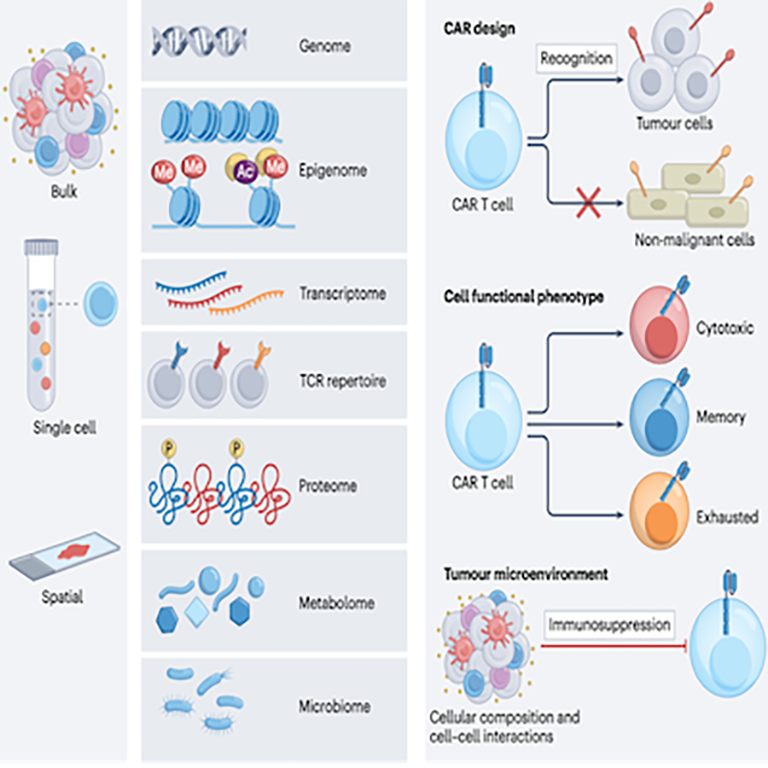
Immunotherapy:
Fully utilizing multi-omics data will provide new insights into the biology of CAR T cell therapy, further accelerate the development of products with improved efficacy and safety profiles, and enable clinicians to better predict and monitor patient responses.
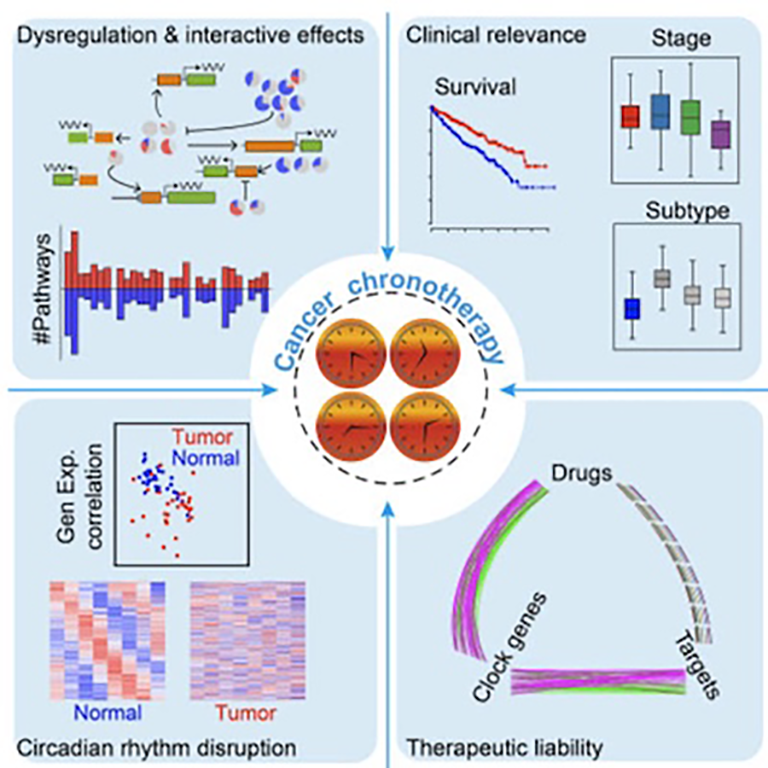
Pan-cancer analyses:
We pioneered a series of pan-cancer analyses to provide clinical insights into cancer therapy, including chronotherapy.
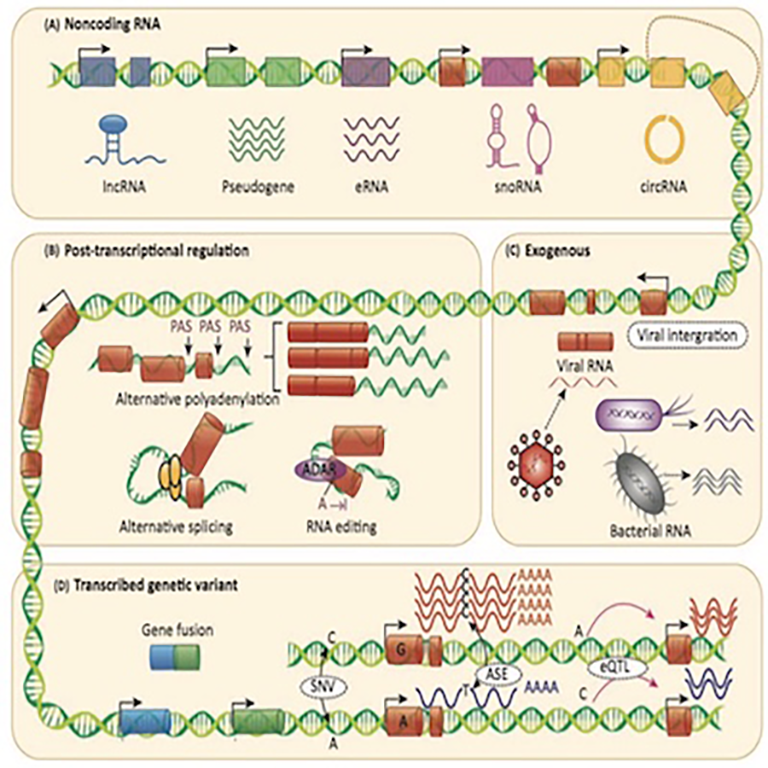
Transcriptomic elements:
We have comprehensive understanding of the molecular mechanisms of novel transcriptomic elements in cancer, including eQTL, snoRNA, APA, circRNA, and eRNA.
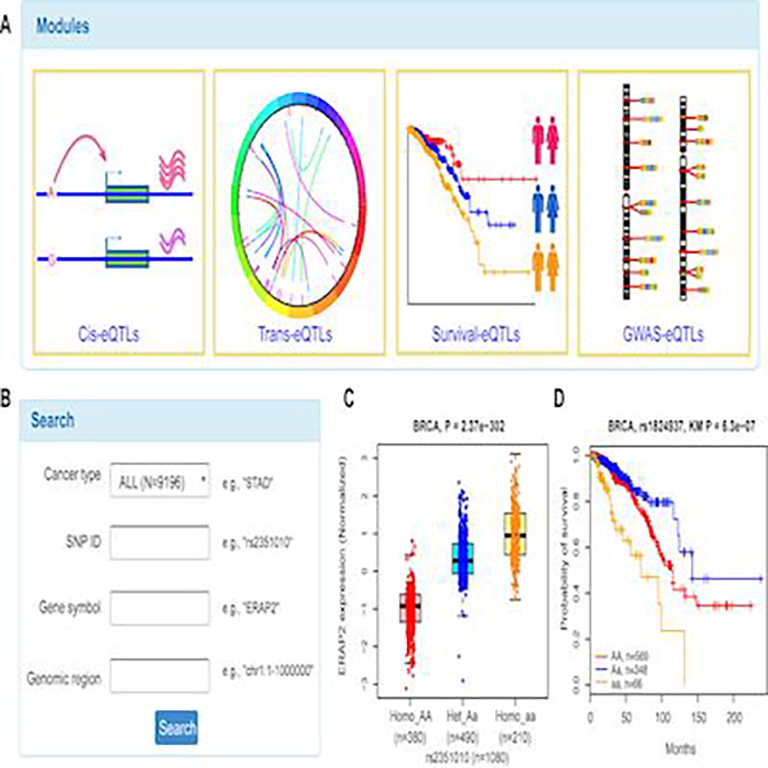
Data resources:
Our group developed several widely used data resources (available below) that provide valuable assets for the research community to further perform functional investigations.
It is challenging to facilitate the utilization of large-scale data by the broad biomedical community without computational background. We are developing further data resources for various types of complex diseases to accelerate investigations in biomedical research.
- eRIC: Database for enhancer RNAs in cancer
- tRIC: Database for tRNAs in cancer
- CircRic: Database for circular RNAs across approximately 1000 cancer cell lines
- APAatlas: Database for Alternative PolyAdenylation atlas in human tissues
- GPSno: Database for the genetic and pharmacogenomic landscape of snoRNAs
- HeRA: Data portal for Human enhancer RNA atlas
- GPEdit: Data portal for genetic and pharmacogenomic landscape of A-to-I RNA editing in cancers
- CSCD2: Integrated interactional database of cancer-specific circular RNAs
- CARTSC: Data portal for chimeric antigen receptor (CAR) target gene toxicity at the single-cell level
- GPIeR: Genetic, pharmacogenomic, and immune landscapes of eRNAs
- IMiT: Comprehensively characterizing the associations between intratumor microbiome with immune checkpoints
- IMCC: Intratumor microbiome associated with clinical characteristics
- GPIP: Genetic, pharmacogenomic, and immune landscapes of cancer proteins
- E3Atlas: Comprehensive characterization of E3 ligases
Liu Y, Yang J, Wang T, et al. Expanding PROTACtable genome universe of E3 ligases. Nature communications 2023, Oct; 16;14(1):6509
Yang J, Chen Y, Jing Y, et al. Advancing CAR T cell therapy through the use of multidimensional omics data. Nature reviews. Clinical oncology 2023, Apr;20(4):211-228
Luo M, Liu Y, Hermida LC, et al. Race is a key determinant of the human intratumor microbiome. Cancer cell 2022, Sep; 12;40(9):901-902
Jing Y, Yang J, Johnson DB, et al. Harnessing big data to characterize immune-related adverse events. Nature reviews. Clinical oncology 2022, Apr;19(4):269-280
Ye Y, Zhang Y, Yang N, et al. Profiling of immune features to predict immunotherapy efficacy. Innovation (Cambridge (Mass.)) 2022, Jan; 25;3(1):100194
Jing Y, Liu Y, Li Q, et al. Expression of chimeric antigen receptor therapy targets detected by single-cell sequencing of normal cells may contribute to off-tumor toxicity. Cancer cell 2021, Dec; 13;39(12):1558-1559
Interested in joining our lab at the Brown Center for Immunotherapy? Don't miss our great opportunities!
Team Science: Meet the Han Lab crew

Yuan Liu, PhD
Research Assistant Professor
PhD, MS, North Dakota State University BS, Michigan State University and Beijing Forestry University
yl218@iu.edu

Jingwen Yang, PhD
Postdoc Fellow
PhD, Fudan University BS, Southern Medical University
jy82@iu.edu

Yamei Chen, PhD
Postdoc Fellow
PhD, Chinese Academy of Medical Sciences and Peking Union Medical College BM, Fujian Medical University
yamchen@iu.edu

Mei Luo, PhD
Postdoc Fellow
Postdoc, Xiangya Hospital of Central South University PhD, Huazhong University of Science and Technology BS, China Three Gorges University
luomei@iu.edu

Chengxuan Chen, PhD
Postdoc Fellow
PhD, Institute of Genetics and Developmental Biology, Chinese Academy of Sciences BS, South China Normal University
cc123@iu.edu

Runhao Wang, PhD
Postdoc Fellow
PhD, North Dakota State University MS, BS, Qingdao Agricultural University
wangrunh@iu.edu

Lifei Ma, PhD
Postdoc Fellow
PhD, Chinese Academy of Medical Sciences and Peking Union Medical College MS, Autonomous University of Barcelona BS, Hebei Agricultural University
TBD@iu.edu
Lab alumni
- Dr. Zhao Zhang, PI at Fudan University
- Dr. Hang Ruan, Professor at Soochow University
- Dr. Shengli Li, PI at Shanghai Jiaotong University
- Dr. Ying Jing, PI at Greater Bay Area Institute of Precision Medicine
- Dr. Youqiong Ye, Professor (PI) at Shanghai Jiaotong University
- Dr. Jing Gong, Professor (PI) at Huazhong Agricultural University




